Part II : How to install NCBI BLAST+ on window 7
|
|
In my previous post, I have discussed that how to download the NCBI BLAST on Window 7 and what is the uses of different software of NCBI local BLAST package. In this Bioinformatic tutorial, I will try to explain that how to use these packages. In NCBI BLAST+, beside the regular BLAST programs like blastn, blastp, there are other several programs for specific purposes.
3. Installation
Notwithstanding the nature of program, you have to always make a database then use the program. As I mentioned previously, your working directory will contain two folders - bin and doc. Folder bin will be your working directory. So open your command prompt and direct it to your working directory i.e. bin
Also Read Part I : How to install NCBI BLAST on window 7
3. Installation
Notwithstanding the nature of program, you have to always make a database then use the program. As I mentioned previously, your working directory will contain two folders - bin and doc. Folder bin will be your working directory. So open your command prompt and direct it to your working directory i.e. bin
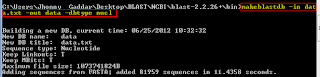
Now, we have to prepare our database. In this example, my database contains nucleotide and stored in data.txt file while my query sequences are stored in query.txt.
NOTE: Please keep in mind that you can know all option for every BLAST programs by typing 'PROGRAM NAME -HELP'. For example, if you want to know the all option for 'blastn -HELP'Since, I am using the nucleotide sequence then I will use blastn options. Lets create database for our database data.txt by typing these command in your command prompt. Depending upon size of yout
database, this process will take few seconds to several minutes. database formatting will results in three file - .nhr, .nin and .nsq file. Now when your database is ready for BLAST, you are ready for blast. You can simply go for blastn following this command
Hope this will help, if you have any problem, then do let me I will try to solve the issue.
Note to me : Use -outfmt 6 to get results in tabular form
|
Related Posts BLAST,
HOW TO,
NCBI
|
Was This Post Useful? Add This To Del.icio.us Share on Facebook StumbleUpon This Add to Technorati Share on Twitter |
Subscribe to:
Post Comments (Atom)







Hi. I am from Universiti Sains Malaysia (USM), Malaysia. I am new in this field. I already follow your guidance and meet an error saying that I didn't meet with Fasta format. Can I know what is fasta format type you used in order to get the blast database result?
ReplyDeleteWelcome Azman. I used typical FASTA formatted nucleotide or amino acid sequences like
Delete>my sequence
AGTCATCGAGTCAGTCAGTCAGTC
Can you share the part of your input file so I can check what is wrong with your file. Thanks
Thank you for your reply.Typically, I have 102540 protein sequences. This is one of them.
ReplyDelete>110597
CCCCOCCCCCCNCNCCCOOCON
Actually, this is my own database. I generated this data from MDDR Database which is one of chemical database and I converted them into amino acid alphabet. I'm done with the transformation process. Now, I want to search the similarity of the transformation MDDR database using Blast. Lastly, I will verified either the E-value and bit-score are equivalent with recall value in chemoinformatics. Basically, I want to use transformation chemical data with bioinformatics searching tool. And after that, I realized that if I use Blast server from NCBI, I will not get the result because NCBI server might not have my database there. Is it my assumption true? That's why when using BLAST, I always keep error like 'testdb.fasta did not meet with FASTA format...' . I hope you can help me.
Are you sure that they are amino acid? As 'o' doesn't stand for any amino acid. You can send me part of your file @ prp291@gmail.com so that I can look into. Thanks
ReplyDelete